The Molecular Architecture of Native BBSome Obtained by an Integrated Structural Approach
Chou H, Apelt L, Farrell DP, White SR, Woodsmith J, Svetlov V, Goldstein JS, Nager AR, Li Z, Muller J, Dollfus H, Nudler E, Stelzl U, DiMaio F, Nachury MV, Walz T(2019) Structure 27: 1384-1394
- PubMed: 31303482
- DOI: https://doi.org/10.1016/j.str.2019.06.006
- Primary Citation of Related Structures:
8ZZI - PubMed Abstract:
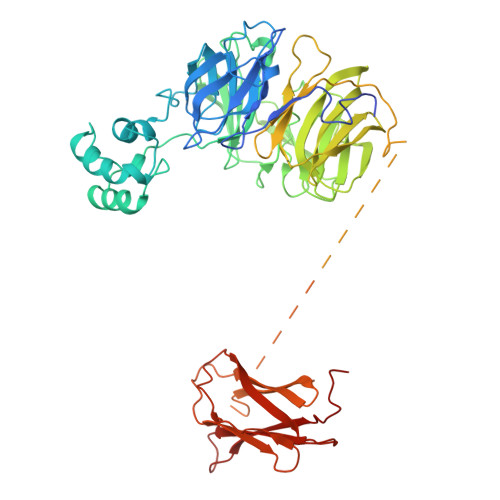
The unique membrane composition of cilia is maintained by a diffusion barrier at the transition zone that is breached when the BBSome escorts signaling receptors out of cilia. Understanding how the BBSome removes proteins from cilia has been hampered by a lack of structural information. Here, we present a nearly complete C¦Á model of BBSome purified from cow retina. The model is based on a single-particle cryo-electron microscopy density map at 4.9-? resolution that was interpreted with the help of comprehensive Rosetta-based structural modeling constrained by crosslinking mass spectrometry data. We find that BBSome subunits have a very high degree of interconnectivity, explaining the obligate nature of the complex. Furthermore, like other coat adaptors, the BBSome exists in an autoinhibited state in solution and must thus undergo a conformational change upon recruitment to membranes by the small GTPase ARL6/BBS3. Our model provides the first detailed view of the machinery enabling ciliary exit.
Organizational Affiliation:
Department of Cell Biology, Harvard Medical School, Boston, MA 02115, USA.